Introduction to FLReconstruct
FLReconstruct's task is to read data from an output file generated by the SuperNEMO simulation or detector, perform reconstruction on each event read, and write the reconstructed data to an output file. It uses a pipeline architecture for event processing, the pipeline is constructed as a sequence of "modules", the sequence of modules being selected by the user (i.e. you) at runtime. Falaise provides a standard set of pipelines and modules, and you can write your own custom pipeline scripts and modules which FLReconstruct can load at runtime via a plugin mechanism.
Here we present a brief overview of running FLReconstruct from the command line using the standard pipeline scripts. The more advanced topics of scripting the pipeline and writing custom modules are covered in the Writing FLReconstruct Pipeline Scripts and Writing FLReconstruct Modules tutorials.
Using FLReconstruct on the Command Line
The flreconstruct program is a command line application just like any other Unix style program (e.g. ls). In the following, we will write commands assuming that flreconstruct is in your path. If it is not, simply use the relative or absolute path to flreconstruct.
You can get help on the options that can be passed to flreconstruct by running it with the -h or --help options, e.g.
The --version option provides detailed information of the current status of the application, including which libraries it uses:
The exact versions you see will depend on the version you are using and the versions of external packages linked.
To examine a data file output by the flsimulate application, you need to supply, at minimum, an input file. By default, this will simply dump information on each event held in the file to standard output (i.e. the current terminal). For example, say we have a file example.brio, then we can dump events in that file by doing:
Quick start
Here we propose to run FLReconstruct with the default setup. Only an input simulation file is set from the command line.
We first generate a single simulated event, using the default simulation setup, and store it in an output file:
Then we process this file with a default pipeline:
The output on the terminal is:
As we can see, the default behaviour of FLReconstruct is to print the contents of the event record(s) in the standard output. We can thus identify the simulated event record with its SD bank (simulated data) which contains the informations about the generated primary event, the original vertex and some collections of truth hits (here in the calo and gg hit categories).
Scripting FLReconstruct
As seen above, if supplied with no other information other than an input file, flreconstruct simply dumps data to standard output. To work with and process the data, a pipeline script must be provided by the user.
Basic Script Syntax
FLreconstruct scripts use the datatools::multi_properties format from Bayeux. The script must begin with a mandatory header:
Comments can be placed at any point in the script. Just prepend a sharp ('#') symbol to any comment line. You may also prepend a comment at the end of a line:
Note that lines starting with ‘’#‘’ are datatools::properties directives, and will be interpreted differently by the parser.
After the header, the script contains sections. A section starts with a section definition line with two identifiers:
- the name of the section,
- the type of the section (in
flreconstructscripts it must be"flreconstruct::section").
The syntax is:
The section body uses the datatools::properties format from Bayeux. After the definition line, an optional description may be provided using the '#@config' directive:
Then comes the section body which consists in a list of parameter setting directives. The format is:
where NAME is the identifier for the parameter, TYPE is its type and VALUE the selected value for this parameter. Some parameters may use an optional DECORATOR which gives additional information about the parameter's type or processing. Example:
How to run a mock calibration on simulated events
When we generate simulated events with FLSimulate, the so-called SD hold the raw simulated calorimeter and tracker hits. In order to be able to reconstruct and analyze the events, we must apply a calibration procedure to transform these "truth" data into the "calibrated" data with the same format as the real data will have.
This calibration procedure consists of determining physics/geometrical observables associated to hits : deposited energy, particle times of flight, hit positions in the geometry model and so on.
The so-called mock calibration derives the calibrated data from the raw simulated data.
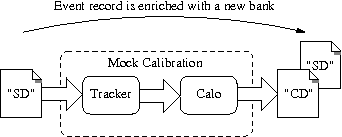
The mock calibration consists of:
- Tracker hits: compute the drift radius and longitudinal position along the anode wire of the Geiger avalanche (and associated uncertainties).
- Calorimeter hits: compute the total energy deposit in the scintillator block and the reference time when the particles first interacted with the block (and associated uncertainties).
A new CD bank holding these data is added in the event record, alongside the raw SD bank.
To see this processing in action, we first create a set of 10 simulated events (using the default simulation setup), using the following simu.conf script for FLSimulate:
and run:
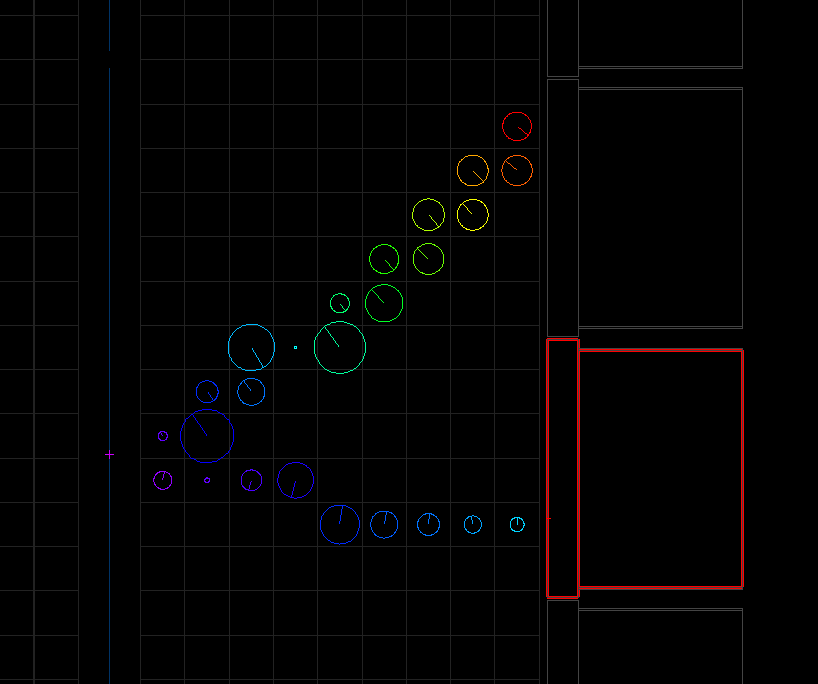
The resulting example.brio file can be opened with the flvisualize application. We can visualize the first event where we will see any raw tracker/calorimeter hits, as well as the position of the decay vertex (cross on the source foil). An example event is shown below, and your view may differ as by default flsimulate will use different seeds on each run, and the styling/colours may also vary between machines:
To process this data in flreconstruct and apply the mock calibration we create the rec.conf script with a custom pipeline inline module made of three consecutive modules. The first one is responsible of the calibration of the tracker hits. The second one is responsible of the calibration of the calorimeter hits. The last one simply prints the content of the events in the standard output:
We use default settings for each of the CalibrateTracker, CalibrateCalorimeters and Dump modules so the corresponding sections are empty.
We then run:
The use of the Dump module prints the events to the terminal. Here is the first event:
We clearly see that a new CD bank (calibrated data) has been added to the event record. It contains two collections of hits, respectively calorimeter and tracker hits.
The example-cd.brio file output by flreconstruct and the pipeline script may be opened in flvisualize as before. Looking at the first event again, we can see additional rendering and information associated to the CD hits (as before, your output will differ due to different random number seeds used in flsimulate):
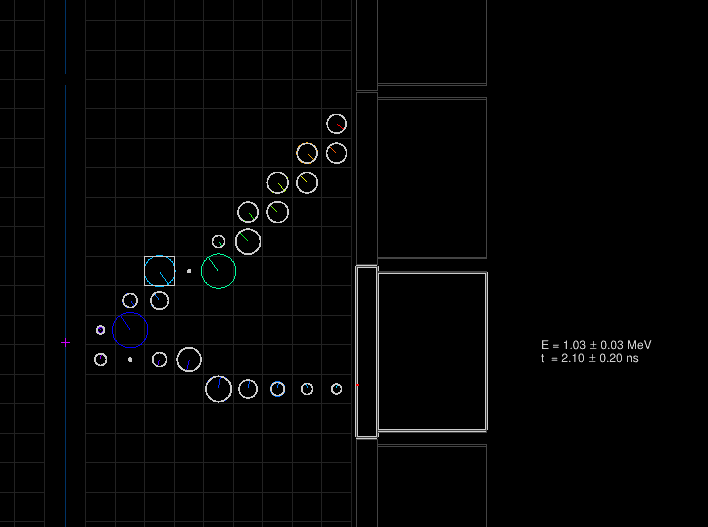
The superimposed white circles on top of the colored ones represent the calibrated tracker hits for which the drift radius and longitudinal position along the anode wire have been computed from the truth hits' timestamps. Here, there is only one calorimeter hit, the total deposited energy in the block and time of entering the scintillator block are also computed (with respect to the arbitrary decay time set by the simulation engine).
How to run a tracking algorithm on simulated events
Now we know how to perform the calibration step, we are interested in the reconstruction of electron tracks in the tracking chamber. For that we need additional processing on top of the CD bank which is now available in the event record.
The reconstruction of tracks associated to charged particles traversing the tracking chamber is a two step procedure:
- first the calibrated Geiger hits are clusterized in tracker clusters, using some special vicinity criteria,
- then a fit is performed on each cluster to compute helix or line segments compatible with the Geiger hits.
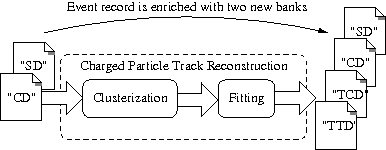
We create a new rec.conf script with a custom pipeline inline module made of five consecutive modules. The first two perform the mock calibration as shown in the previous section. Then we use a new module CATTrackerClusterizer to run the clustering algorithm, followed by the TrackFitting module to run the line/helix fitting on the found clusters. As before, we add a dump module at the end of the pipeline to print each event to standard output.
As we now use modules implemented by dedicated Falaise plugins, we must explicitly provide a flreconstruct.plugins section with the list of plugins that must be dynamically loaded to allow the pipeline to work:
We run:
Now two banks have been added in the event records:
TCD: the tracker clustering data contains the result of the tracker hit clusterization with one or several solutions, each containing a collection of candidate clusters of hit cells. These clusters are used as the input of the track fitting algorithmTTD: the tracker trajectory data contains the result of the tracker fitting with one or several solutions, each containing a collection of candidate fitted tracks (helix or line).
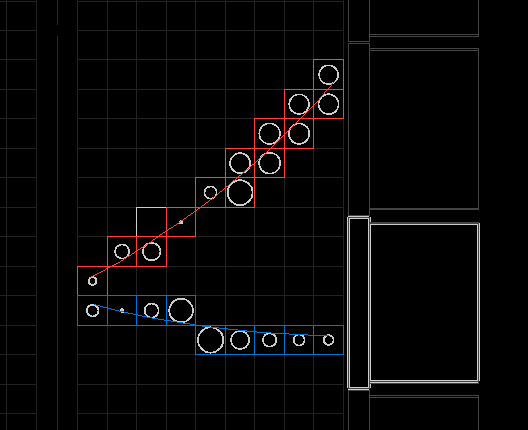
The display shows the best found clusterization/fitting solutions with two clusters of tracker hits (red and blue) and the best associated tracks that have been computed from these clusters. We clearly recognize a two electrons event pattern, but here only one is associated to a calorimeter block.
To do
Document more reconstruction steps:
- charged particle tracking module with extrapolation of vertex, impact points, curvature, track/calorimeter association...
- gamma tracking
- particle identification
Supported sections and parameters in FLReconstruct scripts
The FLReconstruct script contains up to five sections of type flreconstruct::section with the following names:
flreconstruct: this is the system/base section where to set general parameters such as:numberOfEvents: the number of events to be processed from the input (integer, optional, default is:0which means all events will be processed),experimentalSetupUrn: the experimental setup tag (default is:urn:snemo:demonstrator:setup:1.0),
flreconstruct.variantService: this is the variants section where the Bayeux variant service dedicated to the management of variant parameters and configurations is configured. In principle, this section inherits the configuration of the variant service used to generate simulated data.Parameters of interest are:
configUrn: the configuration tag for the variant service associated to the simulation setup (string, optional). If not set, it is automatically resolved from the experiment setup tag.config: the path to the main configuration file for the variant service associated to the simulation setup (string/path, optional). If not set, it is automatically resolved from theconfigUrntag.profileUrn: the configuration tag for the variant profile chosen by the user to perform the simulation (string, optional). If not set, it may be automatically resolved from theconfigUrntag if the variant configuration has a registered default profile.profile: the path to the variant profile chosen by the user to perform the simulation (string/path,optional). If not set, it is automatically resolved from theprofileUrntag or from input metadata.settings: a list of explicit setting for variant parameters chosen by the user to perform the simulation (array of strings, optional). If not set, it is automatically resolved from theprofileUrntag or from input metadata.
flreconstruct.services: this is the services section where explicit configuration for the embedded Bayeux/datatools service manager is defined (by tag or explicit configuration file).Parameters of interest are:
configUrn: the configuration tag for the service manager associated to the data producer setup (string, optional). If not set, it is automatically resolved from the experimental setup tag.config: the path to the main configuration file for the service manager service associated to the reconstrcution setup (string/path, optional). If not set, it is automatically resolved from theconfigUrntag.
flreconstruct.plugins: this is the plugins section where explicit directives are defined to load Falaise plugin libraries which define various types (classes) of processing modules.Parameters of interest are:
plugins: the list of plugins to be loaded.PLUGIN_NAME.directory: the directory from where the plugin dynamic library namedPLUGIN_NAMEshould be loaded (default: "@falaise.plugins:", i.e. the standard location for the installation of Falaise's plugins).
flreconstruct.pipeline: this is the pipeline section where general setup of the reconstruction pipeline is provided.Parameters of interest are:
configUrn: the tag of a registered/official pipeline.config: the main configuration file describing the modules used along the pipeline. If not set, it is automatically resolved from theconfigUrntag.module: the name of the top level pipeline module, chosen from the list of modules defined in the main configuration file (default:"pipeline").
A sample configuration script showing the organisation of the above parameters is shown below. Note that many of the parameters are commented out as they are generally note needed except for advanced use or testing.
The majority of scripts will just use the flreconstruct.plugins and flreconstruct.pipeline sections to select from the set of plugins and pipelines approved by the Calibration and Reconstruction Working Group. Output results from these can be used in the preparation of analyses.
Inline modules
In the case the flreconstruct.pipeline section does not define an explicit pipeline configuration by tag (configUrn) or configuration file path (config), it is possible to provide a list of additional sections which define reconstruction modules. This technique is named the inline pipeline mode. Such a section uses the following format:
where ModuleName is the unique name of the module and ModuleType is the identifier of its registered class type (see below).
This mode should not be used for production runs. It is expected that only official registered pipeline setups are used in such a case, through the configUrn properties in the flreconstruct.pipeline section.
Using Standard Pipelines
FLReconstruct implements a "pipeline" architecture in which events flow through an ordered sequence of "modules". Each module performs a specific task on the event (e.g. count hits), writing its results into the event for further processing by downstream modules. The sequence of modules and their configuration (e.g. algorithm parameters) in the pipeline are constructed via a datatools::multi_properties script. A set of pipeline scripts are supplied with Falaise by the Analysis Board to provide easy to use, validated reconstruction chains. These are located under snemo/demonstrator/reconstruction in the resources tree, with two available:
snemo/demonstrator/reconstruction/official-1.0.0.conf- The "old" pipeline
snemo/demonstrator/reconstruction/official-2.0.0.conf- Current "production" pipeline
snemo/demonstrator/reconstruction/production.pipeline.conf- Synonym for the current production pipeline
The output file generated by flreconstruct using these pipelines may be opened and the results visualized using the `flvisualize` GUI application. To run one of the standard pipelines, it is passed using the -p flag of flreconstruct as follows:
Here @falaise: is a shorthand for "the root directory of Falaise resources" and can be viewed as a "mount point" for standard resource files. The subsequent path is simply the path from that root to the standard reconstruction script.
If the script itself is officially tagged, one can also use directly its tag from the command line:
Please note that this tagging scheme is in flux as it is integrated with Conditions and other databases, and is subject to change.
Writing Reconstruction Results to File
To write flreconstruct's results to file (e.g. for later analysis), the -o option may be used to write processed events to a file supplied as the argument, with the output format chosen based on the file extension.
To output to the BRIO format recognised by flreconstruct use the .brio extension:
The resultant file may be examined directly to see what data has been written by simply passing it back to flreconstruct, e.g.
The resultant file may also be further processed by flreconstruct using a pipeline constructed from your own analysis modules. Please see the document on FLReconstruct Pipeline Output for details of the data structures stored for each processed event. Further documents are available detailing how to write your reconstruction/analysis modules, and how to access event data from modules.
Output to ROOT format is also possible by supplying an output file with the .root extension:
The resultant file contains a single flat TTree structure that may be browsed interactively. Note that currently this format only outputs simulated and calibrated data, with no further reconstruction results.
Using Custom Pipelines
The standard pipelines provided by Falaise are intended to cover a wide range of use cases for standard reconstruction tasks leading to analyses. However, if you need to study improvements to the reconstruction via tuning existing modules or adding new ones then custom pipeline scripts and modules can be used in flreconstruct. The Writing FLReconstruct Pipeline Scripts tutorial covers the syntax and structure of custom pipeline scripts.
 1.8.15
1.8.15